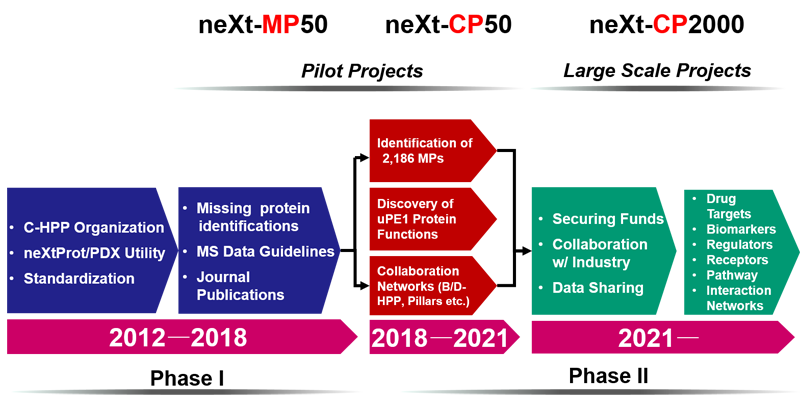
A proposed short- and long-term plan to characterize human dark proteins of unknown functions. The terms, neXt-CP50 and neXt-CP2000 stand for characterizing 50 uPE1, a small set of dark proteins in 3 years (a pilot) and then ~2000 dark proteins (1260 uPE1 plus 677 uMPs) over a longer period of time. Term 'CP' stands for characterization of proteins with unknown functions. (from Paik et al., J. Proteome Res. 2018, 17, 4042−4050)
We believe that high-quality, extensive proteome maps are achievable within a planned 10-year period.
During Phase 1 (6 years), the C-HPP group plans to map all proteins lacking good quality MS evidence,
three major classes of PTMs, one representative alternative splicing transcription (AST) product
[Menon, R. et al., Methods Mol Biol. 2011, 696, 319-26] and one non-synonymous SNP product, and protein
distribution in a major organ/tissue of interest. C-HPP will utilize all high-quality consortium-generated
proteomic datasets for focused analysis on individual chromosomes. In phase II (4 years), identified proteins
will be further characterized and validated at the genomic/transcriptomic and cellular levels in
the 4 selected tissues of interest. C-HPP outputs will also be integrated with all biology/disease-driven
HPP research. We will also provide a correlation of C-HPP and B/D HPP study results with recent SNP and
haplotypic mapping studies. [Im, KM. et al., Hum Genet. 2011, 130, 685-99.]
Table. The proposed two phases of C-HPP and short-term/long-term challenges [Paik. YK., et al., J Proteome Res. 2012, Just Accepted]
aLess well-known: proteins that have only transcriptomic evidence, but not proteomic MS data (constitutes about 6,000 proteins).
bWell-known: proteins that have both transcriptomic and proteomics MS data. The data for proteins under investigation will be integrated into one common C-HPP portal by contributions from each chromosome team.
| Phase | Phase I | Phase II |
|---|---|---|
| Years | 6 | 4 |
| Milestones |
|
|
| Coping with short-term and longer-term challenges | Short-Term | Solutions |
|
|
|
| Longer-Term | Solutions | |
|
|
bWell-known: proteins that have both transcriptomic and proteomics MS data. The data for proteins under investigation will be integrated into one common C-HPP portal by contributions from each chromosome team.





